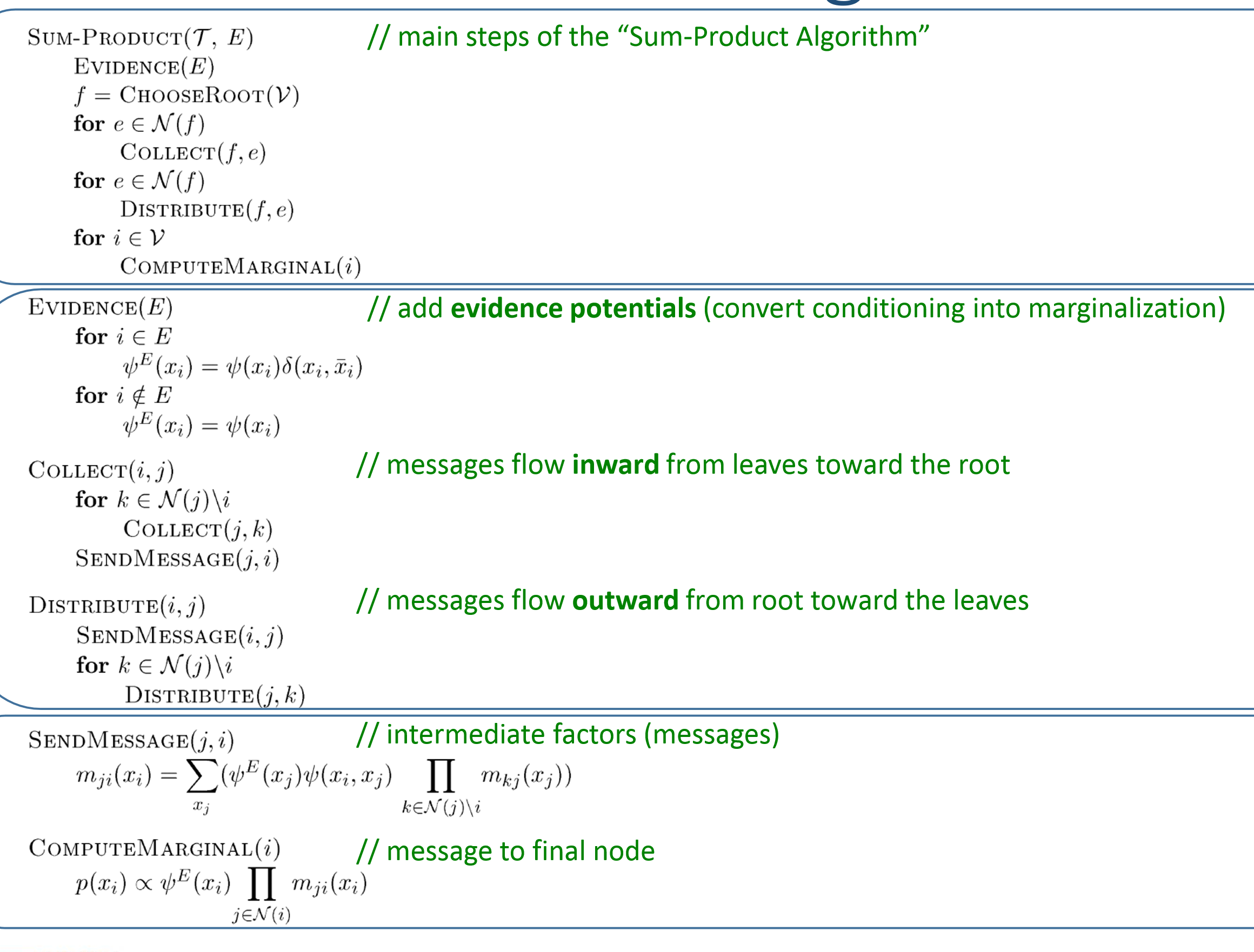
Variable Elimination Algorithm
General probabilistic problem: calculate $p(X_F|X_E)$ for arbitrary disjoint subsets $E$ and $F$: ( $X_F$ are query nodes, $X_E$ are evidence nodes)
$X_R$ are nodes that must be marginalized out of joint probability.
Joint probability table size of $n$ variabels that takes $k$ states:
a naive summation: $O(k^n)$.
a factored form: $O(k^r)$ where $r\ll n$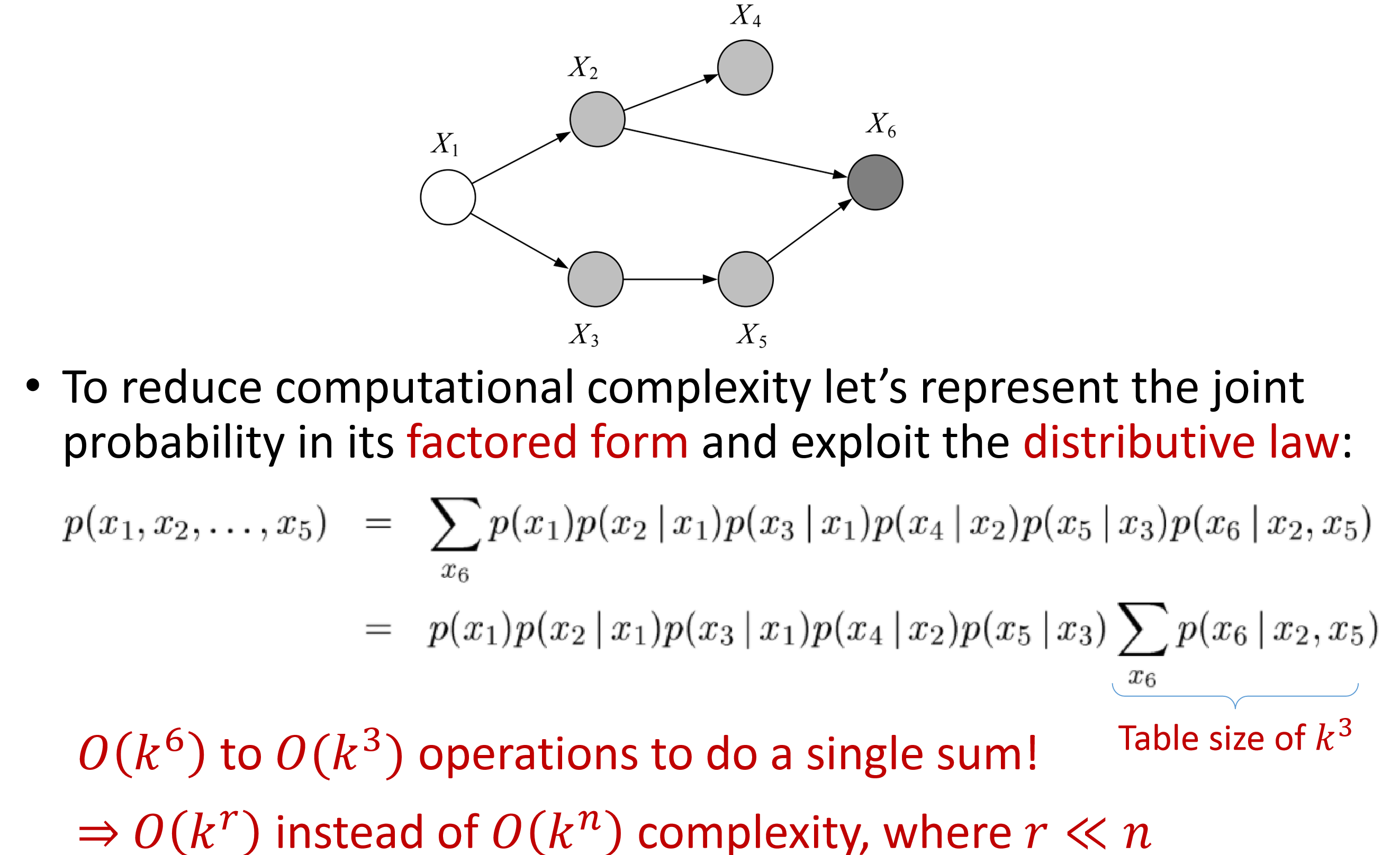
$p(X_6|X_2,X_5)$: 3d table
$p(\bar{X_6}=1|X_2,X_5)$: a 2d slice of the 3d table
Evidence potential:
Turn evaluation into sum:
Which means setting lines of table $p(x_i)$ where $x_i \neq \bar{x_i}$ to 0.
Evidence potential is a trick to simplify the description. In practice we take slices instead of doing summation.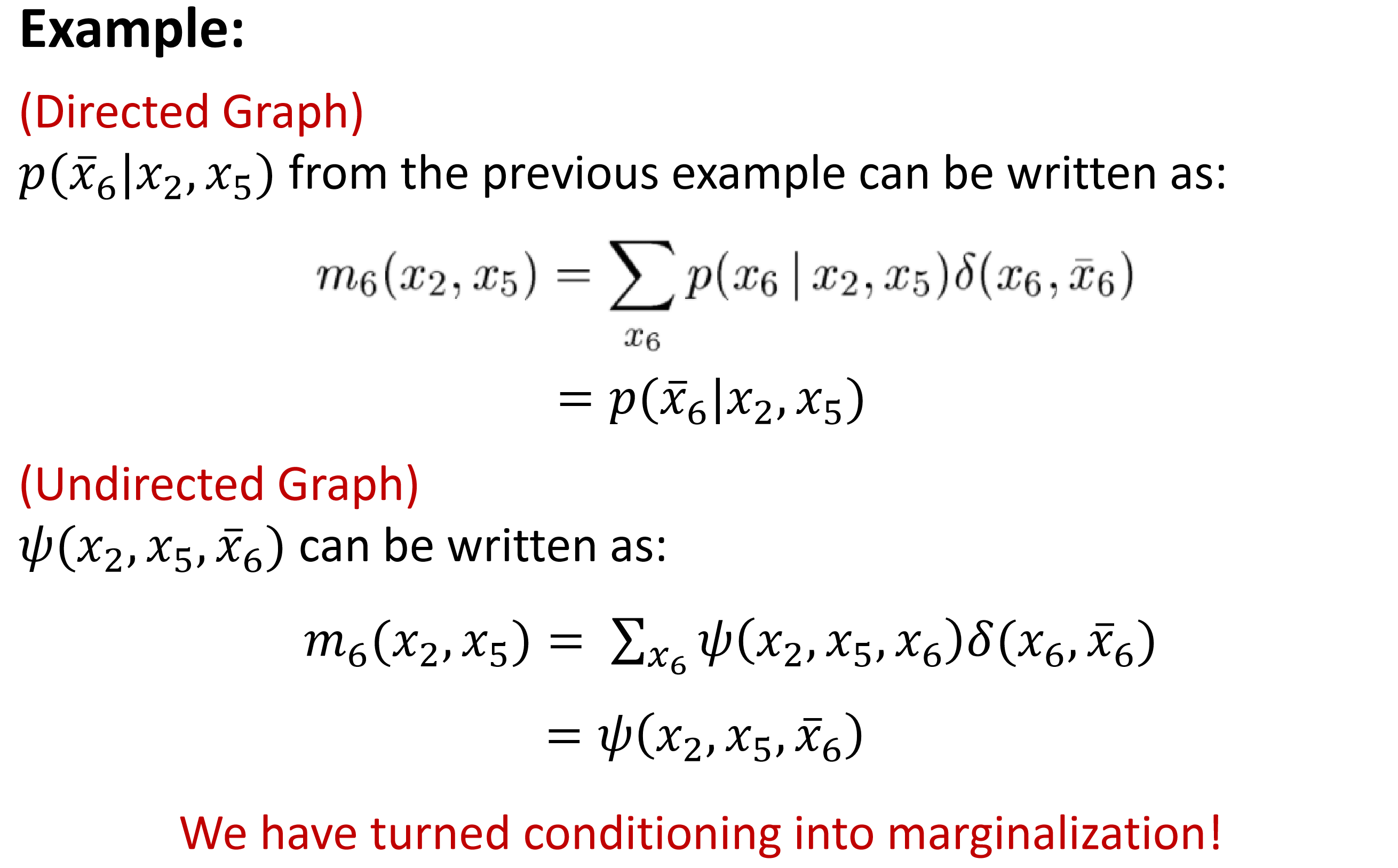
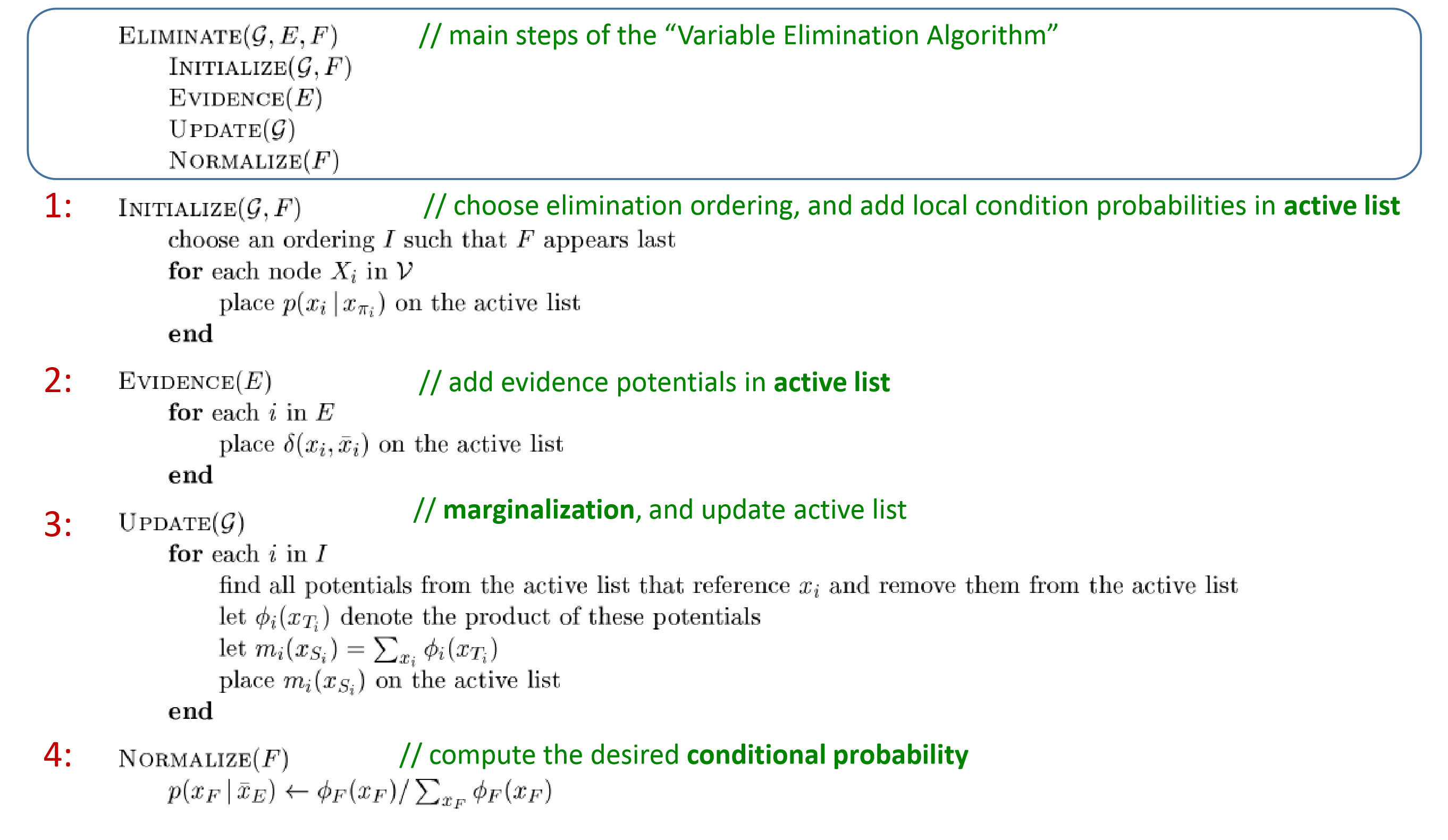
Eliminate(G,E,F):
- Initialize: choose elimination ordering, query nodes appear at the last.
- Evidence: place evidence
- Update: marginalization.
Loop: 1. $\phi_i = $ product of …, which is the unnormalized conditional probability. 2. $m_i(x_{S_i})=\sum_{x_i}\phi_i$, which is the narmalization factor. - Normalize(F)For conditional probability, normalization factor Z cancels.
For a marginal probability, Z cannot be canceled.
Constituted Graph
Eliminate: removing the node from graph and connecting the remaining neighbors, thus creating “elimination cliques”.
Reconstitute: add additional edges during the elimination process to the original graph. (Complexity depends on the choice of elimination order)
Treewidth: one less than the smallest achievable cardinality of the largest elimination clique over all possible elimination ordering.
Computaional Complexity of DGM can be convert to UGM by moralization.
Limitation: re-run elimination algorithm for every new query nodes, since different query nodes require different eliminate order.
Sum-Product Algorithm (Belief Propagation Algorithm)
$ \checkmark$ All single-node marginals
$ \checkmark$ Tree like graphs (undirected tree without loop or can be moralized to such tree)
$ \checkmark$ Using one single run
$ \checkmark$ Cliques are single or pairs of nodes
Convert a directed tree to an undirected one
Define: $\psi(x_r) = p(x_r)$, $\psi(x_i.x_j)=p(x_j|x_i)$, $\psi(x_i)=1\quad\forall i\neq r$, Partition function $Z=1$.
Define local potetials
Construct tree structure
Order: Depth-Fist Traversal.
Take query node $X_f$ as root.
Directing all edges pointing away from $X_f$.
Elimination proceeds inward from leaves with tree width equals to one.
Inward
Send message $(j,i)$:
This message can be re-used.
Compute Marginal:
which is the unnormalized conditional probability.
Message Passing Protocol: a node can send a message to a neihboring node if and only if it has received messages from all its other neighbors.
Two phase approval: inward & outward.